DNA
Genetic Information
-
form?
The genetic information of an organism is stored in the form of nucleic acids.
Nucleic acids, DNA (deoxyribonucleic acid) and RNA (ribonucleic acid), are long linear polymers composed of nucleotide building blocks. Each nucleotide is comprised of a sugar, a phosphate residue, and a nitrogenous bases (a purine or pyrimidine).
DNA is longer than RNA and contains the entire genetic information of an organism encoded in the sequences of the bases. In contrast, RNA only contains a portion of the information and can have completely different functions in the cell.
DNA is structurally characterized by its double helix: two opposite, complementary, nucleic acids strands that spiral around one another. The DNA backbone, with alternatively linked sugar and phosphate residues, is located on the outside. The bases are located inside the helix and form the base pairs adenine and thymine or guanine and cytosine, which are linked by hydrogen bonds.
The human genome comprises 3.2 x 109 base pairs, which are distributed over 23 pairs of chromosomes. Each chromosome is a linear DNA molecule of a certain length. The chromosome is only well visualized under the light microscope during the metaphase of mitosis, as it is maximally condensed during this phase. Chromosomes are present as pairs in most cells of the body. One chromosome in each of the 23 pairs originates from the mother and the other from the father.
Both interrelated chromosomes are termed homologous because they each have a variant of the same gene. Alterations in the number or structure of the chromosomes lead to various conditions, e.g., developmental disorders. Chromosomal assessment with different molecular biology and cytogenetic methods often allows for a clear diagnosis.
Nucleotides
-
Structure
- Nitrogenous base (a purine or pyrimidine)
- A pentose sugar
- Phosphate group
-
Bonds
- Base + sugar → (N-glycosidic bond) → nucleoside + phosphate group at the 3' or 5' C atom of sugar → (ester bond) → nucleotide

-
Derivative
Nucleotide and nucleotide derivatives have important functions in the body.
- Building blocks of nucleic acids
- Source of energy: especially as a universal energy carrier of the cell in the form of ATP, but also GTP
- Signal molecules: especially the second messenger cAMP (cyclic adenosine monophosphate) and cGMP (cyclic guanosine monophosphate) , both phosphoric esters
- Activators for the transfer of groups: Through the potential of forming energy-rich bonds, nucleotides are able to transfer a molecule onto another in biosynthesis, e.g.:
- Regulators: enzyme reactions in signal transduction pathways (e.g., activates GTP G proteins)
- Carrier molecules: e.g., the electron carrier nicotinamide adenine dinucleotide (NAD+) and flavin adenine dinucleotide (FAD) as a component of coenzymes in redox reactions
The energy carrier ATP contains ribose and not deoxyribose as a sugar, and therefore has a 2' OH group!
Nucleobases
-
Nucleobases
- See learning card on purines and pyrimidines for more details.
CUT the PYrimidine; PURine As Gold.
Thymine is found in DNA; uracil is found in RNA.

- Other than uracil, there are many other bases that may be created after the initial nucleic acid chain formation, for example:
A MEAN person **GAG**s a PURring cat!
- Amino acids required for purine synthesis

Phosphate Group
- Phosphate group
- A nucleotide can have one, two, or three phosphate groups (also termed “nucleoside monophosphate”, “diphosphate”, and “triphosphate”, respectively).
- Nucleic acids are composed of nucleoside monophosphates.
- Nucleoside diphosphates and nucleoside triphosphates (e.g., ATP) are found in biochemical processes requiring energy.
Nucleic Acid
-
Nucleic acid sugars
- Structure: The sugar found in nucleic acids is a pentose, which has a five-atom ring. Specifically, the sugar in:
- Pentose binds:
-
overview
-
Dna structure and the human genome
- Overview of DNA structure and packaging
- Double-stranded DNA
- Chromatin
- Chromosomes
- Human genome
- Nuclear genome
- Mitochondrial genome (mitochondrial DNA, mtDNA)
-
RNA classes and their structure
RNAs can be differentiated into various types, which differ in their length, structure, and function. Depending on the type, RNA can be a single-stranded or double-stranded segment.

Laboratory Testing
-
Polymerase chain reaction (PCR)
- what is it?
- common sequence?
- phase
- Uses
- Reverse transcriptase polymerase chain reaction (RT-PCR)
Gene expression and transcription
-
Summary
The genome contains the hereditary information of the structure and function of a cell or organism. This information is stored as a sequence of bases in DNA. A relatively small percentage of DNA codes for proteins and ribonucleic acids (RNAs), while a large amount of the genome is composed of sequences without a clear function. The conversion of the information stored within DNA into a functional molecule, or RNA and proteins, is termed gene expression. Gene expression occurs in two stages: transcription and translation. During transcription, DNA is copied into RNA. RNA is then used to synthesize proteins during translation.
Key enzymes involved in transcription are DNA-dependent RNA polymerases. These enzymes synthesize the RNA molecule based on the genes encoded in DNA, which contain starting sites (promoters) where transcription begins. Transcription factors are required to recognize the promoter. RNA polymerase moves along the template strand of the double-stranded DNA. The strand is synthesized until the end of the DNA segment (termination site) is reached. In eukaryotes, the newly formed primary transcript is further modified to be, for example, available for protein synthesis.
Gene expression is strongly regulated at all levels. Some genes are expressed in all cells and are required as housekeeping genes for basic cellular functions (i.e., constitutive expression). Other genes are only active in certain cells; their expression is regulated by a variety of mechanisms. Genes can undergo activation or silencing, and transcription depends on the presence of specific DNA-binding proteins. The newly formed RNA may also be degraded after transcription by various mechanisms before use in protein synthesis. There are also regulatory mechanisms at a translational level. Although each cell in an organism contains the same DNA, the regulated expression of certain genes causes the cells to specialize and assume different functions, e.g., muscle cells or hepatocytes.
-
Overview
- Gene expression: conversion of genetic information stored in DNA into a functional gene product (RNA and proteins)
- Protein synthesis: process of gene expression (comprised of transcription and translation) as well as post-transcriptional modifications (see the learning card on translation and protein synthesis for more information)
- Central dogma of molecular biology: genetic information always flows in one direction from DNA to RNA to the protein
- DNA → (transcription) → RNA → (translation) → protein
- Exception: retroviruses, which are able to produce DNA from RNA using their own enzyme reverse transcriptase (reverse transcription)
In protein synthesis, DNA is initially transcribed into mRNA (transcription) and mRNA is translated into an amino acid chain (translation)!
Transcription
-
In transcription, DNA serves as a template to produce a complementary RNA molecule. Only a single-strand from the double-stranded DNA (dsDNA) is read.
- DNA segments
- Sense strand: the DNA segment in the double-strand DNA that is complementary to the antisense strand and has an almost identical base sequence to the mRNA that is transcribed along the antisense strand ; The sense strand is not involved in the transcription process.
- Antisense strand: the DNA segment in the double-strand DNA that is used as a template for transcription to produce the complementary mRNA strand.
- Promoter
- Specific DNA sequence located upstream (= in the 5' region) of a gene that regulates transcription
- Contains AT-rich sequences, e.g. TATA box and CAAT box
- Binding site for RNA polymerase II and several other transcription factors at the start of transcription
- Mutations at the site of promoters usually lead to dramatically decreased transcription rate
- Exon-intron structure: eukaryotic genes are composed of alternating coding and noncoding regions
- Substrates: the nucleoside triphosphates ATP, GTP, CTP, and UTP
- Enzymes: RNA polymerases
- General transcription factors: specific helper proteins that help RNA polymerase find and bind to the promoter and initiate RNA synthesis
- DNA segments
-
RNA polymerases and transcription factors
- RNA polymerases
-
Transcription factors
RNA polymerases require helper proteins for promoter recognition of the genes to be transcribed.
- General transcription factors: enable binding of RNA polymerase to the proximal promoter regions by binding of chromosomal DNA to specific base sequences → start of transcription
- Specific transcription factors: modulate transcription by binding to regulatory elements (enhancers, silencers)
- Example: steroid hormone receptors
-
DNA-binding proteins
Proteins, such as transcription factors that bind to DNA, require specific protein domains, also termed structural motifs. These structural motifs usually use either an α-helix or a β sheet to bind to the major groove of DNA. Transcription factors have DNA-binding domains through which they are able to interact with specific DNA segments to perform their function. Numerous structural motifs of DNA-binding domains have been identified. Important examples are the zinc finger domains, leucine zippers, basic helix-loop-helix, and the homeobox.
- Zinc finger
- Leucine zipper
- Characteristics
- Two long α-helices that bind to one another through their hydrophobic regions and form a supercoil
- Because every seventh amino acid residue is leucine and the residues intertwine like a zipper, this structural motif is termed leucine zipper.
- DNA binding: The DNA-binding hydrophilic regions of α-helices contain many basic residues that interact with the major groove of DNA.
- Characteristics
- Basic helix-loop-helix
- Characteristics
- Two polypeptide chains comprising a short and a long α-helix connected by a flexible loop (does not have a secondary structure).
- The two polypeptide chains dimerize via the basic regions of the two α-helices.
- DNA binding: The short basic α-helix interacts with DNA.
- Characteristics
- Homeobox (with helix-turn-helix)
An important structural motif of DNA-binding proteins is an α-helix with many basic amino acid residues!References:[1]
-
Stages of transcription
Transcription is divided into three phases: initiation, elongation, termination.
- Initiation (transcription): the start of transcription by the formation of the initiation complex and unwinding of DNA
- Preinitiation complex (RNA polymerase-promoter closed complex) formation by binding of general transcription factors and RNA polymerase to the promoter region (e.g., TATA box, CAAT box, GC box)
- Formation of a transcription bubble by unwinding the DNA double helix to a single strand with a length of 10–12 bases (open complex)
- Start of RNA synthesis
- Elongation: extension of the RNA strand
- 3'OH group of the growing RNA strand is attached to the α-phosphate group of the next complementary nucleoside triphosphate
- Termination: During termination, polyadenylation starts.
During transcription, base pairing occurs between DNA and RNA. Uracil (instead of thymine) in RNA pairs with adenine in DNA!
RNA and DNA pair in an antiparallel direction. The 5' end of one strand is the 3' end of the other strand and vice versa. In both cases, the base sequences are written in the usual 5'→3' direction!
- Initiation (transcription): the start of transcription by the formation of the initiation complex and unwinding of DNA
-
Post-transcriptional modification (RNA processing)
In eukaryotes, the end-product of transcription is heterogeneous nuclear RNA (hnRNA), which is then transformed into mature mRNA through post-transcriptional modifications in the nucleus. These modifications include capping, polyadenylation, splicing, and RNA editing. mRNA then leaves the nucleus and enters the cytosol
Capping
- Definition: addition of a cap of 7-methylguanosine to the 5' end of hnRNA
- Process
- Cleavage of the 5'-phosphate group by RNA triphosphatase
- Addition of a GMP residue (formed from GTP with cleavage of pyrophosphate) to the 5' diphosphate end of hnRNA by guanylyltransferase
- Methylation of one, two, or three ribosome residues of hnRNA with S-adenosylmethionine (SAM) as a methyl group donor
- Function
- Protects against degradation (through exonucleases )
- Initiation of translation
Polyadenylation
- Definition: addition of a tail of ∼200 adenosine monophosphates (polyadenylate, A) to the 3' end of hnRNA
- Process
- Polyadenylation signal on hnRNA: AAUAAA
- Poly(A) polymerase binds to the cleavage site and adds an ATP-dependent adenosine monophosphate of ∼ 50–250 nucleotides.
- The poly(A) polymerase does not need a template for polyadenylation.
- Function
- ↑ Stability (protects against degradation)
- Initiates translation
Splicing
-
Regulation of transcription
Because transcription and protein synthesis require large amounts of energy, gene expression is strongly regulated. While some genes are continuously transcribed, other genes undergo regulation.
- Prokaryotic gene regulation (operon model)
- Eukaryotic gene regulation
Translation and protein synthesis
Summary
Gene expression is the process by which genetic information flows from DNA to RNA to the protein. The translation of DNA into RNA is termed transcription; protein synthesis from RNA templates is called translation. Details on gene expression and transcription can be found in a separate learning card.
Translation is carried out by ribosomes, which are large molecular complexes of ribosomal RNA (rRNA) and proteins. Ribosomes bind to RNA templates, also termed messenger RNA (mRNA), and catalyze the formation of a polypeptide based on this template. In the process, a charged transfer RNA (tRNA) recognizes a nucleotide triplet of mRNA that matches a specific amino acid (AA). The new AA is then linked to the next AA of the growing polypeptide on the ribosome. Translation ends once a specific nucleotide sequence of the mRNA is reached (a stop codon). The ribosome subsequently dissociates and the mRNA and newly synthesized protein are released.
Before proteins are functional, a proper shape and destination are both necessary. Proteins begin to fold into their three-dimensional structure during translation according to the AA sequence and local chemical forces and reactions. Various specialized proteins (folding catalysts, chaperones) also help the newly formed proteins to fold properly and reach their correct destinations (e.g., cytosol, organelles, extracellular matrix) via protein modifications. The translation rate of proteins is adjusted to the current conditions of the cell and bodily demands, and is affected by the presence or absence of certain nutrients.
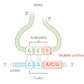
Genetic code
The genetic code
- Description: relationship between the DNA (or mRNA) nucleotide sequence and the respective amino acid sequence of a protein
- Codons
- Codon: sequence of three nucleotides (a triplet) of mRNA that codes for a specific AA
- Anticodon: sequence of three nucleotides in tRNA that is complementary to the codon on mRNA
- The codon and anticodon are always paired in an antiparallel manner.
- There are 64 combinations of codons: 61 for 20 AAs (including the start codon), and 3 for stop codons
- Notable codons
- Start codon: AUG (located at the 5' end of mRNA) initiates translation of the mRNA
- Codes for methionine-tRNA (each polypeptide initially starts with methionine)
- Stop codons: UGA, UAG, or UAA end translation
- Start codon: AUG (located at the 5' end of mRNA) initiates translation of the mRNA
- Features
- Unambiguity (genetic code): codon is specific to only 1 AA.
- Commaless, non-overlapping (genetic code): Each codon of the mRNA is translated in the 5' to 3' direction continuously in an open reading frame (ORF), beginning with the start codon and ending with the stop codon.
- Exceptions: some viruses
- Redundancy, degeneracy (genetic code): The genetic code is redundant, meaning most AAs are encoded by > 1 codon.
- Exceptions: methionine and tryptophan
- Reduces the extent of damage caused by DNA mutations.
- Due to tRNA wobble: Some tRNA molecules are able to recognize multiple codons through a certain degree of flexibility (wobble) between the pairing of the third nucleotide of the codon with the first nucleotide of the anticodon.
-
The wobble position is at the 5' end of the anticodon (3 position of the codon), which usually contains inosine instead of the usual nucleotides
rd
- Hypoxanthine pairs unspecifically with uracil, adenine, and cytosine at the 3' end of the codon.
- Inosine forms wobble base pairs.
-
- Universality (genetic code): Almost all organisms use the same genetic code (has not evolved).
- Exceptions: mitochondria, Mycoplasma, some yeasts, archaebacteria
To help remember the stop codons: UAA → U Are Away, UAG → U Are Gone, UGA → U Go Away

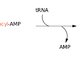
tRNA charging
- Description: binding of tRNA to its corresponding AA
- Reaction mechanism: The charging of tRNA in the cytosol occurs in two steps and is catalyzed by aminoacyl tRNA synthetases.
- Mischarged tRNA:
- AAs are checked before binding tRNA, but if an incorrect AA binds the tRNA, the bond is hydrolyzed
- Any mischarged tRNA inserts the wrong AA into the polypeptide, which is why AA-tRNA synthetase is important for accurate AA selection.
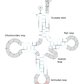
Translation process
Translation occurs in three phases in a functional ribosome: initiation, elongation, and termination. It requires mRNA, tRNA, and rRNA (see RNA section from nucleic acids: DNA and RNA).
Ribosome binding sites
- See “Ribosomes” in the cell for more details.
- Ribosome is composed of rRNA and proteins that form subunits:
- Small subunit (40S subunit in eukaryotes, 30S subunit in prokaryotes)
- Large subunit (60S subunit in eukaryotes, 50S subunit in prokaryotes)
- Binding site for mRNA is on the small ribosomal subunit.
- Both ribosomal subunits jointly form the binding sites for tRNA.
- Aminoacyl site (A site)
- Peptidyl site (P site)
- Exit site (E site)
- Because of the length of most mRNA, more than one ribosome can bind them, allowing synthesis of multiple polypeptides at once = polysome.
Eukaryotes have Even-numbered ribosomal subunits (40S + 60S → 80S); PrOkaryotes have Odd-numbered ribosomal subunits (30S + 50S → 70S).
For binding sites, think of an “APE” party: 1. A site → Arrival with Aminoacyl-tRNA 2. P site → Growing (GTP) Party of Peptides 3. E site → party Ends and is EmptytRNA Exits; Growing stands for GTP as energy source
1. Initiation
- Description: assembly of functional ribosomes with the help of initiation factors (IFs) and recognition of the start codon (AUG) on the mature mRNA by the initiator methionyl-tRNA (met-tRNA)
- Process
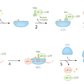
- Initiator met-tRNA, eukaryotic IF2 (eIF2), and GTP bind to the small ribosomal subunit to form a preinitiation complex.
- eIF2: a small G protein
- Binds initiator met-tRNA and forms the final initiation complex by hydrolyzing GTP to GDP
- Reconverted to the GTP-bound form by the guanine nucleotide exchange factor eIF2B
- eIF2: a small G protein
- mRNA is recognized by eIF4 and binds to the preinitiation complex. eIF4 recognizes mRNA:
- Usually at the 5' cap in eukaryotes
- Sometimes at an internal ribosome entry site (IRES)
- A site of mRNA that allows translation initiation without a 5' cap
- Most commonly located in the 5' UTR (especially of RNA viruses like poliovirus), but can be located at many sites in the mRNA and also occurs in eukaryotes
- Initiator met-tRNA recognizes the start codon (typically the first AUG triplet after the 5' cap of the mRNA) and binds the P site.
- GTP hydrolysis provides energy for the release of eIF2, allowing the large and small ribosomal subunits to assemble into a functional ribosome (the final initiation complex; 80S in eukaryotes, 70S in prokaryotes).
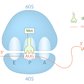
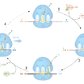
2. Elongation
- Directions of processes
- Process
- Initiator met-tRNA is located at the P-site or another previously matching aminoacyl-tRNA is bound there.
- An aminoacyl-tRNA complex with eukaryotic elongation factor 1 (eEF1) hydrolyzes GTP, thereby releasing eEF1 and GDP and providing the energy for aminoacyl-tRNA to bind the A site (anticodon matches the codon of the mRNA).
- The polypeptide is elongated by the stepwise addition of AAs via peptide bonds between the AAs bound to the A-sites and P-site (via tRNA).
- Bond created by a dehydration reaction catalyzed by a peptidyl transferase that is intrinsic to the rRNA (“ribozyme”) of the large complex
- Ribosomal translocation
- The ribosome moves one triplet along the mRNA in the 3' direction.
- Energy is derived from GTP hydrolysis that is catalyzed by eEF2.
- After translocation, the tRNA that was in the A-site is now in the P-site, and the tRNA that was in the P-site is now in the E-site.
- The unloaded tRNA is released from the E-site.
- The ribosome moves one triplet along the mRNA in the 3' direction.

3. Termination
- A release factor recognizes the stop codon and hydrolytically cleaves the peptidyl tRNA bonds (requires GTP) → release of the protein
ATP for Activating (charging) tRNA; GTP for tRNA Gripping and Going through the ribosome (translocation) for Growing a polypeptide
Protein folding and misfolding
Protein folding
-
Description: The process by which a protein goes from an unfolded native state to form a three-dimensional structure via progressive stabilization of the intermediate states until the most favorable energy level is achieved.
- Correct protein folding begins during translation and is required for a protein to perform its function within the cell or organism.
-
Intrinsic factors
-
The spatial structure is specified in the AA sequence of the protein (see protein structure for more details).
-
Largely driven by hydrophobic interactions, Van der Waals forces, H-bonds, salt bridges, disulfide bonds
-
-
Regulatory proteins: proteins that help other proteins to form their native structure
- Chaperone proteins
- Protein complexes that prevent protein aggregation during synthesis (thus prevent making proteins nonfunctional) and permit refolding of misfolded proteins in a protected environment
- Assist in transporting (precursor) proteins
- ATP is consumed in the process.
- Examples: heat shock proteins (e.g., Hsp70, Hsp60, Hsp90) prevent denaturation or misfolding at high temperatures or when under chemical stress
- Folding catalysts: enzymes that accelerate rate-limiting steps during protein folding
- Protein disulfide-isomerase: Catalyzes the formation of thermodynamically favorable disulfide bonds within proteins (in proteins that possess more than two cysteine residues), if less energetically favorable disulfide bonds are formed.
- Prolyl isomerase: Assists in finding the energetically favorable conformation of a peptide bond with the AA proline.
- Chaperone proteins
Protein misfolding
- Description: lack of folding or non-native protein folding as a result of denaturing factors
- Triggers
- Stress: e.g., oxidative conditions
- Increased body temperature (fever)
- Mutations: e.g., chloride channel misfolding in cystic fibrosis; misfolded hemoglobin in sickle cell anemia; amyloidosis
- Mechanisms of misfolding
- Hydrophobic amino acid residues on the surface of a misfolded protein tend to aggregate with other hydrophobic surfaces/proteins.
- Accumulation of proteins rich in β-sheets can lead to fibrillation.
- Intracellular reaction: Misfolded proteins are identified and either rescued by chaperones or are ubiquitinated and labeled for proteasomal degradation (see protein degradation).
- Consequences of misfolding
- In some disorders, the protective mechanism fails.
- Misfolded proteins form insoluble aggregates and fibrils, which leads to cell damage and death (e.g., Parkinson disease and Alzheimer disease).
- See protein degradation in the learning card on “Proteins and peptides” for examples of associated conditions.
Post-translational modification
Many proteins require specific covalent alterations (co- or post-translational modifications) in addition to correct folding to function properly. Examples include glycosylation, lipid anchors, phosphorylation, acetylation, ubiquitination, ADP-ribosylation, biotinylation, carboxylation, methylation and hydroxylation.
Protein glycosylation
- Enzymatic attachment of a carbohydrate to specific AA side chains of proteins via N-glycosidic (N-linked glycosylation) or O-glycosidic bonds (O-linked glycosylation), forming a glycoprotein
- Glycoproteins are usually cell membrane, lysosomal, or secretory proteins (e.g., serum proteins such as erythropoietin).
N-linked glycosylation, the attachment of sugar to the asparagine residue of proteins, begins in the rough ER!Enzymatic glycosylation should not be mistaken with nonenzymatic glycation. In glycation, aldoses (e.g., glucose) spontaneously bind to the amino groups of proteins and may influence their function! (A classic example is HbA1c, whose function is unaffected by glycation.)
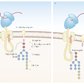

Lipid anchors
- Description: Most membrane proteins interact with lipid membranes via hydrophobic side chains (e.g., valine or leucine residues). However, lipid-anchored proteins are modified to covalently bind lipid anchors.
- Types
- Acylation: linkage with long chain fatty acids, e.g., palmitic acid
- Isoprenylation: linkage of a cysteine side chain of the protein with polyisoprene via a thioester bond, such as in:
- Farnesylation: linkage with a farnesyl residue (three isoprene units, total of 15 C atoms)
- Geranylgeranylation: linkage with a geranylgeranyl residue (four isoprene units, total of 20 C atoms)
- GPI anchor: linkage with glycosylphosphatidylinositol (GPI), a glycolipid
Reversible covalent alterations
Enzymatic reversible protein modification alters the protein's spatial structure (conformation), thereby allowing its activity to be regulated. For example, a protein may interact with other proteins and/or become recognizable as a substrate. Reversible protein modification essentially allows the protein to be switched on or off.
-
Phosphorylation: attachment of phosphate residues to the hydroxyl group (OH group) of serine or threonine or of tyrosine residues by kinases
- Example: regulation of glycogen phosphorylase in glycogen metabolism via phosphorylation
-
Acetylation: linkage with a CO-CHgroup by acetyltransferases
3
- Example: regulation of DNA condensation and with it transcription via acetylation of histone proteins
-
Ubiquitination: attachment of ubiquitin (a small protein) to the ε-amino group of lysine residues in proteins, especially in proteins to be degraded
- Example: cell cycle regulation via ubiquitination and degradation of cyclins
-
SUMOylation: attachment of a small ubiquitin-like modifier (SUMO) protein to lysine residues (similar to ubiquitination)
- Example: Many regulators of the cell cycle are regulated via SUMOylation.
-
ADP-ribosylation: transfer of an ADP-ribose residue from NAD+ by ADP-ribosyltransferase
- Example: histone modification by poly ADP-ribosylation
-
Biotinylation: attachment of biotin
- Example: pyruvate carboxylase uses a biotin cofactor to catalyze the ATP-dependent carboxylation of pyruvate to oxaloacetate
-
Carboxylation: attachment of a carboxylic acid group (R–COOH)
-
Example: vitamin K-dependent carboxylation is involved in liver synthesis of clotting factors that bind Ca
2+
-
-
Hydroxylation: attachment of hydroxy groups (-OH), typically to proline and sometimes lysine (requires vitamin C)
- Example: important for collagen synthesis to ensure crosslinking outside the cell (which explains why vitamin C deficiency leads to scurvy)
-
Methylation: attachment of methyl groups. Methylation of DNA or histones usually results in gene suppression.
Protein trimming
- Description: the excision of N- or C-terminal propeptides to create a mature protein (from an inactive state)
- Examples:
Protein sorting
Protein transportation
A protein's intended final destination depends on its signal sequence (if it has one) at the N-terminus and determines if translation is concluded on free ribosomes or ribosomes on the rough ER.
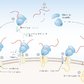
Translocation of ribosomes on the rough ER
Proteins that leave the cell (secretory proteins), as well as membrane and lysosomal proteins, are initially synthesized on free ribosomes of the cytosol. However, their synthesis is paused shortly after starting and the ribosome is transported to the cytosolic side of the rough ER. Protein synthesis then recommences and the protein is directly synthesized into the ER lumen.
Mechanism
- Initiation of translation on the free ribosomes in the cytosol
- If a signal sequence (specific amino acid sequence of 9–12 amino acids) is synthesized, it is bound to a signal recognition particle (SRP, a ribonucleoprotein):
- SRP induces a pause in translation and transports the ribosome with the peptide chain across the ER membrane.
- SRP facilitates binding of the ribosome with the signal peptide to the SRP receptor on the ER membrane.
- SRP and the SRP receptor are both bound to GTP, which is hydrolyzed to GDP. → SRP is released and can bind to a new signal sequence.
- The ribosome is transferred to a translocon, a protein-lined channel composed of a complex of proteins spanning the ER membrane, with opening of the translocon channel. This translocon is termed the sec61 channel.
- Translation resumes and the protein is synthesized in the ER lumen.
- The signal sequence is cut off from the growing protein by a signal peptidase.
- After termination of translation, the ribosome is released into the cytosol.
- The translocon channel closes and the synthesized protein is left in the ER.
- During translation, the protein is folded into its native conformation.
If the SRP is absent or dysfunctional, there will be an accumulation of proteins in the cytosol of the cell!
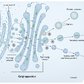
Other processes
- Protein modification in the ER
- N-linked glycosylation
- Formation of disulfide bonds
- Protein distribution
- Proteins destined for the cell membrane are directly anchored in the ER membrane.
- Process: Hydrophobic segments are usually present in the amino acid sequence, which are then integrated into the ER membrane.
- Soluble proteins for lysosomes or proteins destined to leave the cell via exocytosis remain following completion of translation until they are transported into the ER lumen.
- All newly synthesized proteins in the ER are transported via the Golgi network to their target destination by transport vesicles.
- Lysosomal proteins contain a mannose 6-phosphate residue, which is recognized and bound by the membrane-bound mannose 6-phosphate receptor in the trans-Golgi network and transports the proteins to lysosomes in vesicles.
- Soluble proteins destined to remain in the ER are labeled by the signal sequence KDEL (encodes the letters of the amino acids lysine-aspartate-glutamate-leucine) on their C-terminus.
- Only properly folded proteins can leave the ER and travel to their final destination.
- Proteins destined for the cell membrane are directly anchored in the ER membrane.
Translational regulation
- Main component: initiation phase of translation (regulation via initiation factors)
- Regulation mechanisms
- Decreased formation of the ternary complex of eIF2, GTP, and initiating tRNA through phosphorylation of the initiation factor eIF2
- Phosphorylated eIF2 in the GDP-bound form can no longer be converted to its active GTP-bound form. The ternary complex necessary for initiating translation can no longer be formed and the translation rate is reduced.
- Example: synthesis of globin in red blood cells
- If there are insufficient levels of heme available to be incorporated in the globin protein, globin synthesis is inhibited by phosphorylation of eIF2.
- Example: synthesis of globin in red blood cells
- Phosphorylated eIF2 in the GDP-bound form can no longer be converted to its active GTP-bound form. The ternary complex necessary for initiating translation can no longer be formed and the translation rate is reduced.
- Regulation of the cap recognition process by eIF4
- Decreased formation of the ternary complex of eIF2, GTP, and initiating tRNA through phosphorylation of the initiation factor eIF2



Tidak ada komentar: